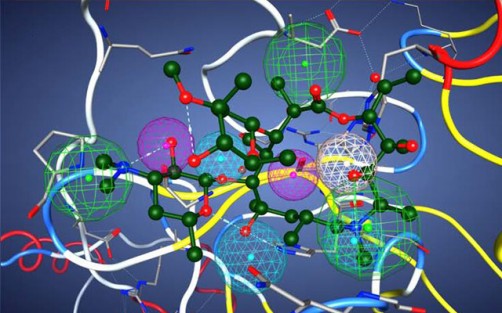
This image illustrates azithromycin in the pharmacophore model of a potential protease inhibitor. The model contains nine functional centers: two donors, two acceptors, one donor/acceptor center, and three hydrophobic centers. Courtesy of V. Kouznetsova, I. Tsigelny, D. Huang, San Diego Supercomputer Center/UC San Diego
San Diego Supercomputer Center (SDSC) researchers Valentina Kouznetsova and Igor Tsigelny recently created a pharmacophore model and conducted data mining of the conformational database of FDA-approved drugs that identifies 64 compounds as potential inhibitors of the COVID-19 protease. Among the selected compounds are two HIV protease inhibitors, two hepatitis C protease inhibitors, and three drugs that have already shown positive results in testing with COVID-19.
The conformations of these compounds underwent three-dimensional fingerprint similarity clusterization. The researchers also conducted docking of possible conformers of these drugs to the binding pocket of protease and then conducted the same docking of random compounds.
“This cocktail of inhibitors could help save thousands of lives,” said Tsigelny. “We have now published our findings with Chemrxiv so that we can move on with proper testing that would next allow this concept to be utilized during the current pandemic crisis.”
“There are several lines of defense when a virus like this attacks the organism and one of them is the siege of the fortress of the cell with a virus – there are inhibitors and antibodies that prevent COVID-19 to bind a receptor ACE2 to infuse to the human cell,” explained Kouznetsova. “To help visualize this, it is as if people on the fortress moor are throwing cobblestones and pouring hot tar on the heads of enemies. The second line of defense is when the enemies arrive and hand-to-hand combat begins. This latter line is what protease inhibitors do – they don’t allow the virus to produce needed viral proteins for a further attack.”
Tsigelny further explained that the main point of this study is elucidation of FDA-approved drugs that can be immediately tried and used on patients – and avenue supported by other COVID-19 research.
“The possibility that existing drugs could be useful immediately for countering this infection is a very important topic,” said Roberto Burioni, a professor of microbiology and virology at Milan's Vita-Salute San Raffaele University and author of the recently released Virus: The Great Challenge: From Coronavirus to the Plague: How Science Can Save Mankind. “To fight an epidemic you need speed and strategy. The later the reaction, the more likely the defeat.”
In addition to their positions at SDSC, Tsigelny, and Kouznetsova are also affiliated with UC San Diego’s Moores Cancer Center. Tsigelny is also affiliated with the UC San Diego Department of Neurosciences and CureMatch, Inc. High school student David Huang, a participant in SDSC’s Research Experience for High School Students (REHS) program, conducted the computational docking.
Source: University of California San Diego
Be the first to comment on "Researchers Develop Potential COVID-19 Protease Inhibitors"