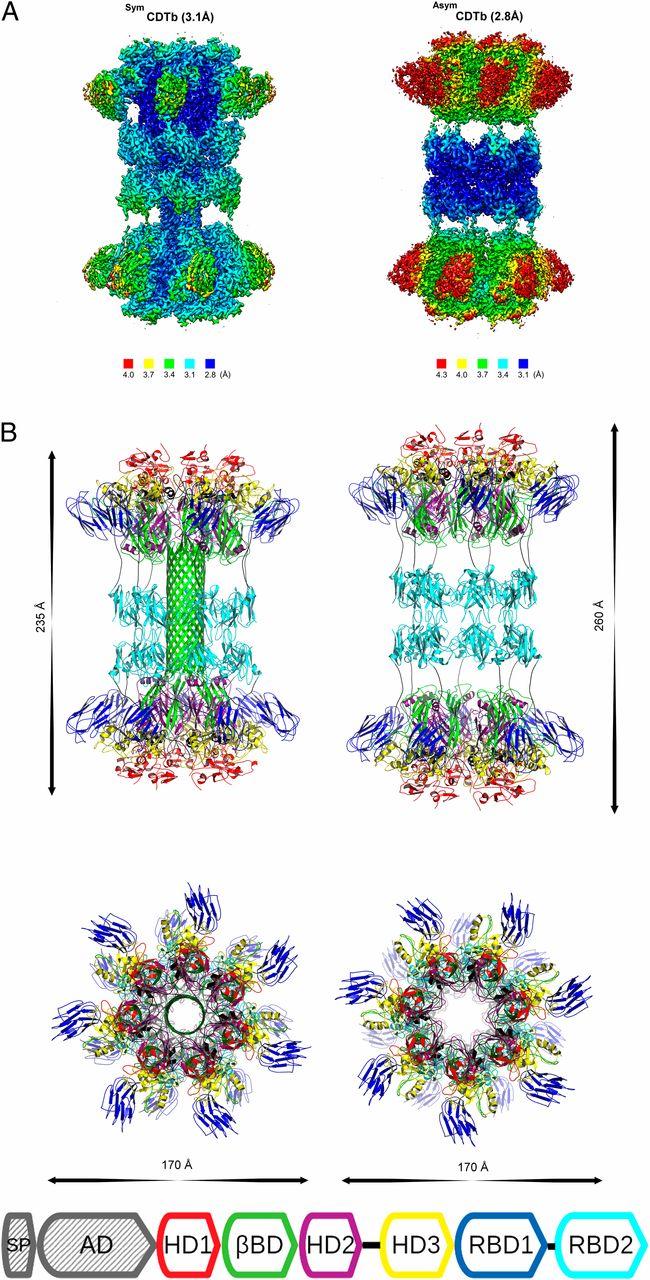
(A) Local resolution in structures of AsymCDTb and SymCDTb conformations. (B) Overall structure of the activated CDTb tetradecamer in AsymCDTb and SymCDTb conformations. Courtesy of PNAS
Researchers from the University of Maryland School of Medicine and their colleagues have identified the structure of the most lethal toxin produced by certain strains of Clostridium difficile bacteria, a potentially deadly infection associated with the use of antibiotics. The finding, published today in the Proceedings of the National Academy of Sciences journal, used cryo electron microscopy, X-ray crystallography and other biophysical methods to identify the microscopic structures of the bacteria. The researchers mapped out the delivery and binding components of the toxin, which could pave the way for new drugs to neutralize it.
"We identified two structures that help explain the molecular underpinnings of C. difficile toxicity," said study co-author David Weber, PhD, a professor of biochemistry and Molecular Biology and director of the Center for Biomolecular Therapeutics at UMSOM. "These structures will be important for targeting this human pathogen using structure-based therapeutic design methods."
Certain C. difficile infections are notoriously difficult to treat and often arise from antibiotic therapy given to fight other infections, which wipes out the beneficial bacteria that normally keep C. difficile bacteria in check. More than 500,000 cases occur annually in the United States, according to the Centers for Disease Control and Prevention, resulting in an estimated 15,000 deaths, usually from strains that are resistant to antibiotic treatment.
There is currently no drug approved by the Food and Drug Administration that targets the C. difficile toxin (CDT) or "binary toxin".
The work was done in conjunction with researchers at the City University of New York, City College of New York, the National Institute of Standards and Merck & Co.
With the mapping of the structures of one of the binary toxins, called CDTb, the researchers next aim to answer important new questions regarding the molecular mechanism of the C. difficile toxin, which can also benefit drug discovery. For example, they will investigate how the active binary toxin complex assembles and dissociates, how it binds to a healthy cell in a patient's gastrointestinal tract and how it enters the cell.
This research was funded by the National Institutes of Health, National Science Foundation Division of Chemistry, and the U.S. Department of Health and Human Services.
Be the first to comment on "Researchers Identify Key Structure of C. difficile That Could Lead to Future Treatments"