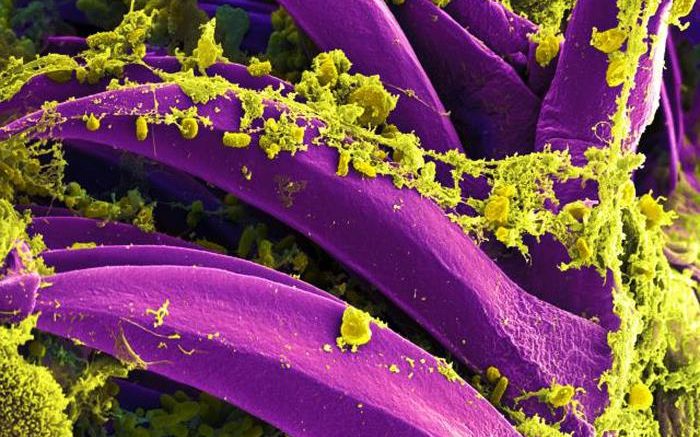
Produced by the National Institute of Allergy and Infectious Diseases (NIAID), this digitally colorized scanning electron microscopic (SEM) image depicts a number of yellow-colored, Yersinia pestis bacteria, that had gathered on the proventricular spines of a Xenopsylla cheopis flea. These spines line the interior of the proventriculus, a part of the flea’s digestive system. The Y. pestis bacterium is the pathogen that causes bubonic plague. Courtesy of National Institute of Allergy and Infectious Diseases (NIAID)
The Yersinia genus covers a vast range of bacteria that are distinguished by criteria such as whether or not they are able to cause disease (their pathogenicity). For instance, Yersinia pestis causes plague, while the bacteria Yersinia enterocolitica and Yersinia pseudotuberculosis are responsible for bowel diseases. Researchers at the Institut Pasteur have developed a new genomic analysis method for classifying and identifying all Yersinia strains and estimating their pathogenicity.
The Yersinia genus, part of the family Enterobacteriaceae, currently includes 19 species and three pathogens affecting humans: the plague pathogen Yersinia pestis; and the foodborne pathogens Yersinia enterocolitica and Yersinia pseudotuberculosis, which cause enteric yersiniosis, a disease that is transmissible by food.
Yersinia enterocolitica is the third biggest cause of bacterial diarrhea in temperate and cold countries after Salmonella and Campylobacter. In France, infections mostly occur in isolated cases or affect small groups.
"Previously, strains were identified using biochemical methods, which sometimes lack precision. They rely on metabolic reactions, leading to misidentification in the event of atypical reactions," says Cyril Savin, deputy director of the National Reference Center (CNR) for Plague and other Yersinia Infections, hosted by the Institut Pasteur.
In such circumstances, an identification strategy applicable to all species of Yersinia, including those that have not yet been described, is a useful asset. Scientists in the Institut Pasteur Biodiversity and Epidemiology of Bacterial Pathogens Unit therefore joined forces with the CNR and the Bioinformatics and Biostatistics Hub in the Department of Computational Biology to develop a genome-based method of identifying strains of the Yersinia genus (a genome includes all the genes contained in DNA).
"We developed a method for analyzing the genome sequence, enabling it to be translated quickly and comprehensibly," explains Sylvain Brisse, head of the Biodiversity and Epidemiology of Bacterial Pathogens Unit. "When we scanned the sequence of numerous genes in the Yersinia genome for each strain, we realized that each bacterium has its own unique genetic profile. The method involves transforming this genetic profile into a sort of standardized 'barcode'."
By applying this method to the Yersinia genus, over 3,000 barcodes were identified, some of which revealed new species. "Our work culminated in a standardized language enabling all labs to recognize these codes," continues Brisse. To disseminate the method, public access was provided to a database of "barcodes" (genomic profiles) and associated identification details, enabling labs throughout the world to identify Yersinia strains using their own genomic sequences.
Using an automated classification algorithm, each genomic profile is linked to its species and genetic lineage.
"Our comparison of the genetic profiles of Yersinia strains revealed an unexpected level of biodiversity including several new and previously unknown species. Strains can now be identified extremely reliably based on their genomic profile", says Alexis Criscuolo, a bioinformatician in the Department of Computational Biology. "Using this method, we can also more accurately define the pathogenicity of strains sent to us at the CNR. Approximately 33 percent of them are not pathogenic!" emphasizes Savin.
Accurate identification of strains is, in fact, essential since pathogenicity differs among Yersinia strains: "It enables more effective patient monitoring and can also be used to guide the rollout of public health initiatives," explains Javier Pizarro-Cerdá, head of the Yersinia Research Unit at the Institut Pasteur.
The analysis of microbial strain diversity plays a key role in addressing infectious diseases. As part of its 2019-2023 Strategic Plan, the Institut Pasteur is seeking to expand its expertise of microbial strain taxonomy (a branch of biology aimed at classifying living organisms into groups known as taxons with a view to identifying them). This genomic taxonomy project is part of a broader program. In order to increase the impact of research on health, the Institut Pasteur is engaged in a project aimed at organizing, better characterizing, and promoting the Institut Pasteur's biological collections in partnership with the National Reference Centers (CNR) and member institutes of the Institut Pasteur International Network. This vision links in with the concept of global health based on the idea that health should be addressed at global level and also that human, animal, and environmental health, which are interdependent and form a whole (concept of "one health") should be addressed jointly. Within this framework, other genomic taxonomy projects have already been successfully completed, enabling monitoring, classification and decoding of strains of other bacteria researched at the Institut Pasteur such as Listeria, Leptospira, Klebsiella, and the rarer Elizabethkingia.
Unrestricted sharing of baseline genomic profiles and their associated barcodes will facilitate research on genetic relationships between strains, enabling the detection of outbreaks and improved understanding of how strains are transmitted at global level and between the environment, animals, food, and humans.
Source: Institut Pasteur
Be the first to comment on "Yersinia is a Novel Genomic Tool for Identifying Strains"