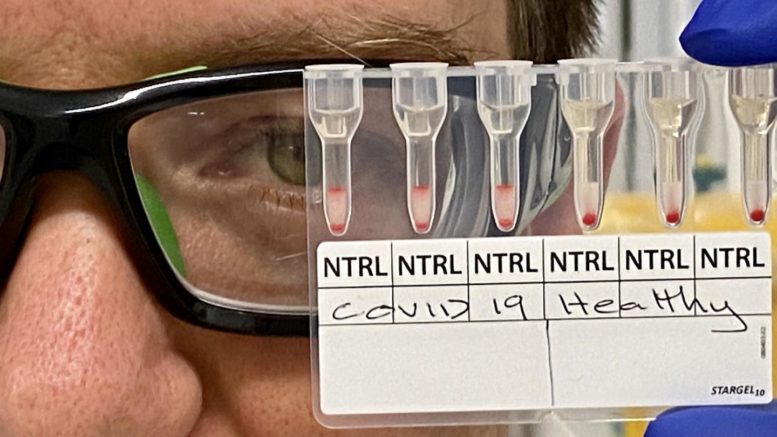
Dr. Simon Corrie with COVID-19 positive and negative blood samples. Courtesy of Monash University
World-first research by Monash University in Australia has been able to detect positive COVID-19 cases using blood samples in about 20 minutes, and identify whether someone has contracted the virus.
In a discovery that could advance the worldwide effort to limit the community spread of COVID-19 through robust contact tracing, researchers were able to identify recent COVID-19 cases using 25 microlitres of plasma from blood samples.
The research team, led by BioPRIA and Monash University's Chemical Engineering Department, including researchers from the ARC Centre of Excellence in Convergent BioNano Science and Technology (CBNS), developed a simple agglutination assay - an analysis to determine the presence and amount of a substance in blood - to detect the presence of antibodies raised in response to the SARS-CoV-2 infection.
Positive COVID-19 cases caused an agglutination or a clustering of red blood cells, which was easily identifiable to the naked eye. Researchers were able to retrieve positive or negative readings in about 20 minutes.
While the current swab / PCR tests are used to identify people who are currently positive with COVID-19, the agglutination assay can determine whether someone had been recently infected once the infection is resolved - and could potentially be used to detect antibodies raised in response to vaccination to aid clinical trials.
Using a simple lab setup, this discovery could see medical practitioners across the world testing up to 200 blood samples an hour. At some hospitals with high-grade diagnostic machines, more than 700 blood samples could be tested hourly - about 16,800 each day.
Study findings could help high-risk countries with population screening, case identification, contact tracing, confirming vaccine efficacy during clinical trials, and vaccine distribution.
This world-first research was published July 18 in the journal ACS Sensors.
A patent for the innovation has been filed and researchers are seeking commercial and government support to upscale production.
Dr. Simon Corrie, professor Gil Garnier and professor Mark Banaszak Holl (BioPRIA and Chemical Engineering, Monash University), and associate professor Timothy Scott (BioPRIA, Chemical Engineering and Materials Science and Engineering, Monash University) led the study, with initial funding provided by the Chemical Engineering Department and the Monash Centre to Impact Anti-microbial Resistance.
Corrie, senior lecturer in chemical engineering at Monash University and Chief Investigator in the CBNS, said the findings were exciting for governments and health care teams across the world in the race to stop the spread of COVID-19. He said this practice has the potential to become upscaled immediately for serological testing.
"Detection of antibodies in patient plasma or serum involves pipetting a mixture of reagent red blood cells (RRBCs) and antibody-containing serum/plasma onto a gel card containing separation media, incubating the card for 5-15 minutes, and using a centrifuge to separate agglutinated cells from free cells," Corrie said.
"This simple assay, based on commonly used blood typing infrastructure and already manufactured at scale, can be rolled out rapidly across Australia and beyond. This test can be used in any lab that has blood typing infrastructure, which is extremely common across the world."
Researchers collaborated with clinicians at Monash Health to collect blood samples from people recently infected with COVID-19, as well as samples from healthy individuals sourced before the pandemic emerged.
Tests on 10 clinical blood samples involved incubating patient plasma or serum with red blood cells previously coated with short peptides representing pieces of the SARS-CoV-2 virus.
If the patient sample contained antibodies against SARS-CoV-2, these antibodies would bind to peptides and result in aggregation of the red blood cells. Researchers then used gel cards to separate aggregated cells from free cells, in order to see a line of aggregated cells indicating a positive response. In negative samples, no aggregates in the gel cards were observed.
"We found that by producing bioconjugates of anti-D-IgG and peptides from SARS-CoV-2 spike protein, and immobilizing these to RRBCs, selective agglutination in gel cards was observed in the plasma collected from patients recently infected with SARS-CoV-2 in comparison to healthy plasma and negative controls," Professor Gil Garnier, Director of BioPRIA, said.
"Importantly, negative control reactions involving either SARS-CoV-2-negative samples, or RRBCs and SARS-CoV-2-positive samples without bioconjugates, all revealed no agglutination behavior."
Professor Banaszak Holl, head of chemical engineering at Monash University, commended the work of talented PhD students in BioPRIA and Chemical Engineering who paused their projects to help deliver this game changing COVID-19 test.
"This simple, rapid, and easily scalable approach has immediate application in SARS-CoV-2 serological testing, and is a useful platform for assay development beyond the COVID-19 pandemic. We are indebted to the work of our PhD students in bringing this to life," Holl said.
"Funding is required in order to perform full clinical evaluation across many samples and sites. With commercial support, we can begin to manufacture and roll out this assay to the communities that need it. This can take as little as six months depending on the support we receive."
COVID-19 has caused a worldwide viral pandemic, contributing to nearly 600,000 deaths and more than 13.9 million cases reported internationally (figures dated July 17, 2020).
Source: Monash University
Be the first to comment on "Breakthrough Blood Test Detects Positive COVID-19 Result in 20 Minutes"