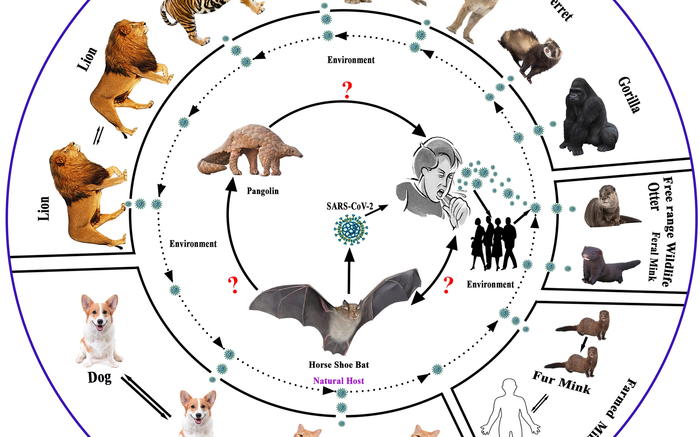
Possible evolution and transmission pathways of SARS-CoV-2 and its related virus at animal-human- ecosystem interface. Courtesy of Islam et al.
A genetic and geographic analysis of variants of SARS-CoV-2—the virus that causes COVID-19—and related viruses in humans and animals may provide evidence of interspecies transmission worldwide. Dr. Ariful Islam, of EcoHealth Alliance and the Bangladesh Institute of Epidemiology, Disease Control and Research, and colleagues present these findings in the open-access journal PLOS ONE on Dec. 15, 2021.
Prior research has suggested that SARS-CoV-2 might have originated through genetic changes that occurred among closely related viruses in horseshoe bats. Reports also suggest that the virus can spread from humans to domesticated and wild animals (known as spillback). However, much remains to be learned about the epidemiology, evolutionary dynamics and genetic relationships between SARS-CoV-2 and related viruses in animals around the world.
To provide new insights, Islam and colleagues conducted a comprehensive analysis of the genome sequences of SARS-CoV-2-related viruses found in bats and pangolins, and of the SARS-CoV-2 strains that have been found in a variety of animals across the globe, including dogs, cats and lions.
Genetic analysis showed that SARS-CoV-2 strains found in animals are closely related to strains found in humans in the same geographic regions -- including Germany, France, Spain, and Denmark -- which evidences the theory that human to animal transmission has occurred worldwide. In future, if the virus becomes established in animal hosts, that animal species might act as a SARS-CoV-2 reservoir.
The researchers also quantified the degree to which genetic mutations associated with key SARS-CoV-2 subtypes and variants are found in animal species in different regions. Multiple emerging variants of concern, such as the Alpha, Delta and Mu-variants, were detected across many countries in species including dogs, gorillas, lions, and cats, and these variants showed notable genetic similarity with human SARS-CoV-2 sequences. For example, cats and American mink were frequently infected with one subtype of virus known as the GR clade (31.6% and 49.7% of samples respectively), which is also often seen in humans, supporting the likelihood of interspecies transmission. However, most dogs were affected by a different subtype, clade O (66.7%), which the authors suggest indicates a particular affinity of clade O for dogs. The SARS-CoV-2 Alpha variant comprised just 2.6% of cat and 4.8% of dog samples, but 66.7% of gorilla and 77.3% of lion samples.
Their genetic analysis revealed very high similarity between SARS-CoV-2 and related viruses found in multiple horseshoe bat species, as well as high similarity with related viruses in the Malayan pangolin. The analysis supports the hypothesis that SARS-CoV-2 originated from closely related bat viruses that genetically recombined with each other, and that the virus also passed through pangolins.
Animals appear to be susceptible to SARS-CoV-2 and may contribute to its spread. Thus, on the basis of their findings, the researchers call for continued genetic monitoring of SARS-CoV-2 in animals. They also call for prevention of contact between infected humans and animals, as well as vaccination of pets, zoo animals, and farm animals. The researchers advocate for surveillance at the human-animal interface to detect and prevent emergence of future viruses.
The authors add, “Spillover and spillback of SARS-CoV-2 has been proved, and the mutant strain of the virus is still dominating. It is our responsibility not to destroy the natural habitats of wildlife. As bats play an important role in maintaining a healthy ecosystem, we need to work on how to live safely with bats. We should not wait until the next pandemic, and we need to apply the One Health approach to secure a healthier future for the world.”
Source: PLoS ONE