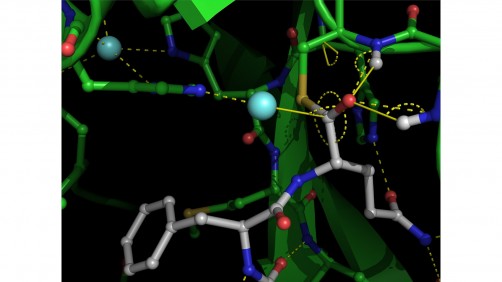
X-ray crystallographic structure of SARS-CoV-2 main protease. Courtesy of University of British Columbia.
The COVID-19 vaccines currently rolling out are providing hope that the spread of the disease can be halted. But infection rates are still high, and for those who contract COVID-19, the search for effective treatments remains important.
Researchers examining the atomic structure of SARS-CoV-2, the virus that causes COVID-19, have made a landmark discovery that could contribute critical information to the design of safe and effective antiviral drugs in the fight against the virus.
Using a powerful X-ray beam to study SARS-CoV-2 proteins in crystallized form, a team from the University of British Columbia (UBC) has observed — for the first time ever — the virus’s main protease, an important enzyme of the virus, performing its function.
This widely pursued antiviral target is a central enzyme that allows the virus to cut up large proteins called polyproteins into smaller functional units, a process necessary for the virus to be replicated and infect other human cells.
“What we’ve captured at high resolution is one of the important steps in that process that has never been visualized before in any viral protease of this class,” said Natalie Strynadka, the UBC biochemistry professor who led the research team with colleague Mark Paetzel.
The research was published in Nature Communications.
The breakthrough was made possible by the Advanced Photon Source (APS), a U.S. Department of Energy (DOE) Office of Science User Facility at DOE’s Argonne National Laboratory. The APS produces X-rays that are roughly a billion times brighter than those used by doctors and dentists, allowing researchers to examine the structure of the coronavirus protease in very fine detail at the atomic level.
Data was captured at the General Medical Sciences and Cancer Institutes Structural Biology Facility at beamline 23-ID-B at the APS.
The newly uncovered information may be of particular interest to scientists worldwide who are racing to develop antiviral treatments for COVID-19. If the main protease is inhibited by a small molecule drug, the polyproteins won’t be clipped into functional pieces, effectively blocking viral replication and subsequent transmission.
“We now have a much better blueprint of these mechanistic structures that will inform making the best inhibitor possible,” Strynadka said. “Better knowing the structure as we now do helps guide drug research, narrowing the field of potential targets instead of having to screen billions of potential molecules.”
Michael Becker, a protein crystallographer with Argonne’s X-ray Science Division, said Strynadka’s research stands out because the team was focused on understanding the mechanism of the protease.
“This understanding will improve everyone else’s work in designing drugs,” Becker said. “Because the more deeply you understand how something works, the better the chance you have of controlling or stopping it.”
Remote access capabilities at Argonne made it possible for the researchers in British Columbia to collect data in real time and to manipulate the APS beamline located about 2,200 miles away in Illinois. UBC team members Jaeyong Lee and Liam Worrall shipped crystals of the SARS-CoV-2 main protease preserved in liquid nitrogen from Canada to Argonne. Workers at the APS were on hand to answer questions, ensure the working order of the equipment, and load the samples.
“The remote interface is fantastic. It’s almost like being there,” Strynadka said. “We’re very thankful for the use of the APS. Canada does have a national synchrotron facility, but it currently doesn’t have the same capability as the APS, which is a very high-level facility with micro-focused beams. Understanding enzymes goes hand in hand with understanding their atomic structures — and the higher resolution the better, because subtle differences can affect the interpretation. We wanted the best data possible, so we went to the APS."
Source: Argonne National Laboratory
Be the first to comment on "New Data About SARS-CoV-2’s Functions Could Aid in Treatment Designs"