While the Centers for Disease Control and Prevention (CDC) coordinates several outbreak and clinical surveillance systems for norovirus, norovirus is strongly under-reported due to individuals not seeking care or not being tested. As a result, norovirus surveillance using case reports and syndromic detection often lags rather than leads outbreaks. Digital epidemiology sources such as search-term data may be more immediate, but can be affected by behavior and media patterns.
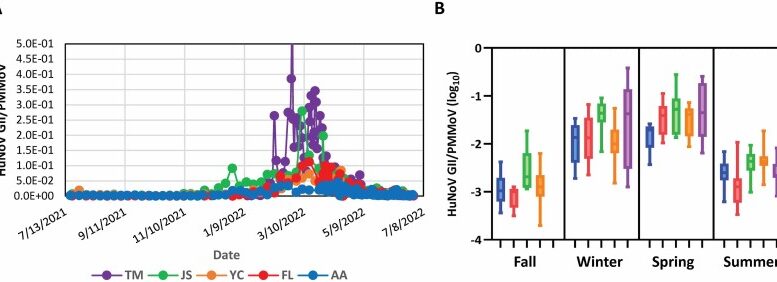
Wastewater monitoring can potentially provide a comprehensive and consistent data stream that can help to triangulate across these different data sets. To assess the timeliness of norovirus wastewater testing compared with syndromic, outbreak and search term trend data for norovirus, Ammerman, et al. (2024) quantified human norovirus GII in composite influent samples from five wastewater treatment plants (WWTPs) using reverse transcription-digital droplet PCR and correlated wastewater levels to syndromic, outbreak, and search term trend data. Wastewater human norovirus (HuNoV) GII RNA levels were comparable across all WWTPs after fecal content normalization using Pepper mild mottle virus (PMMoV). HuNoV GII wastewater values typically led syndromic, outbreak, and search term trend data.
The best correlations between data sources were observed when the wastewater sewershed population had high overlap with the population included by other monitoring methods. The increased specificity and earlier detection of HuNoV GII using wastewater compared to other data, and the ability to make this data available to healthcare, public health, and the public in a timely manner, suggests that wastewater measurements of HuNoV GII will enhance existing public health surveillance efforts of norovirus.
Reference: Ammerman ML, Mullapudi S, Gilbert J, Figueroa K, de Paula Nogueira Cruz F, Bakker KM, et al. (2024) Norovirus GII wastewater monitoring for epidemiological surveillance. PLOS Water 3(1): e0000198. https://doi.org/10.1371/journal.pwat.0000198
Source: PLOS Water