Much remains to be discovered on how the HIV-1 virus infects our cells. Scientists know that it slips past the defenses of our immune system, entering white blood cells to deliver its genetic payload and hijack the cell’s transcription machinery that in turn cranks out copies of viral RNA and new HIV-1 viruses. But many of the details remain hazy.
A major experimental discovery made in 2021 shined some light on the mystery and found that the viral capsid, a protein envelope protecting its RNA genome, remains intact all the way into the nucleus of the target cell. Ultimately, the capsid has to stay stable long enough to take its deadly genetic cargo into the nucleus of the cell. But in the end, it has to break apart to release its genetic material. What scientists don’t yet know is how and why the HIV-1 viral capsid can become unstable.
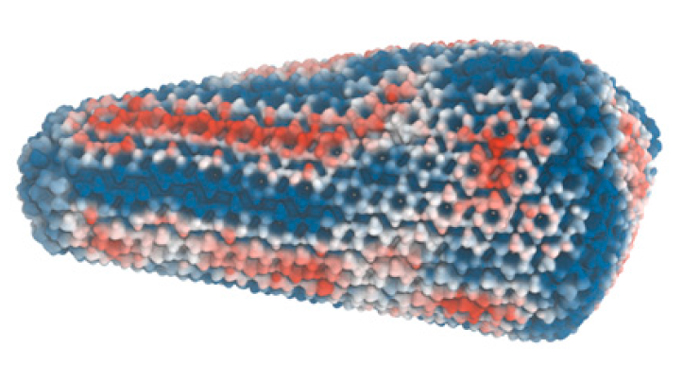
The Frontera supercomputer at the Texas Advanced Computing Center at The University of Texas at Austin has furthered scientists’ understanding of how the HIV-1 virus infects and helped generate the first realistic simulations of its capsid, complete with its proteins, water, genetic material, and a key cofactor called IP6 recently discovered to stabilize and help form the capsid.
“The vulnerabilities in the armor of the HIV-1 virus capsid were revealed by these very big simulations and the analysis we did,” said Gregory Voth, the Haig P. Papazian Distinguished Service Professor at the University of Chicago. Voth is the lead author on the HIV-1 capsid study published in March 2022 in the Proceedings of the National Academy of Sciences.
Voth and colleagues started with cryo-electron tomography data from actual viruses obtained by the lab of co-author John Briggs, Department of Cell and Virus Structure, Max Planck Institute of Biochemistry. Using the experimental data, they developed an all-atom molecular dynamics simulation of the HIV-1 capsid that approached a whopping 100 million atoms.
The images in the study show striations on the capsid, which indicate stress-strain. They pin-point where the protein lattice is compressed or expanded and is undergoing torturous strain, which tells scientists that the stress-strain is not perfectly distributed.
“That's very critical because we were able to correlate those patterns of how the lattice is strained with how the capsids actually break apart,” Voth remarked. The stress-strain striations should be vulnerable to the pressure generated within the HIV-1 viral capsid as it starts undergoing reverse transcription and begins producing DNA.
The stress-strain patterns correlate well, concluded the authors, with how the capsid breaks apart through additional cryo-electron tomography experiments by one of the study collaborators Owen Pornillos, of the Department of Molecular Physiology and Biological Physics, University of Virginia.
“It’s the most realistic simulation study of HIV capsid to date,” Voth said. “We could also see that the proteins that pack into this virus capsid have somewhat different conformations than are seen in simpler crystal structures or in vitro reconstitutions.”
Voth pointed to prior work published in 2017 in Nature by Juan R. Perilla, University of Delaware, and the late Klaus Schulten, University of Illinois at Urbana-Champaign and co-workers. Those authors developed the first model of the HIV-1 capsid on the Blue Waters supercomputer. However, as pioneering as it was at the time, it lacked the inner genetic material, the IP6 co-factor, and wasn’t built from cryo-electron tomography data of actual virus capsids.
The new model built in all of these missing elements. “Our work is a big step forward in the realistic modeling, which I think leads us to a greater understanding of this capsid,” Voth said.
One approach taken by the drug company Gilead in producing the HIV-1 treatment lenacapavir takes knowledge about the HIV-1 viral capsid to make it more brittle, which interferes with its critical break-up stage prior to unleashing its genetic material.
“That's what we're doing now,” Voth said, “is to study, given the heterogeneous nature of this virus capsid, can we understand how drugs interact with it, and can we design new drugs?”
What’s more, drug designers are interested in the potential of a one-two punch pharmaceutical, where one drug molecule binds to the capsid protein lattice, which in turn helps a different drug molecule bind.
Said Voth, “Supercomputers combined with the methods we developed have helped reveal essential elements of the HIV-1 virus that are experimentally extremely difficult to probe at present. I don't think we could have easily done those simulations anywhere else but on Frontera. It’s a very valuable resource for us.”
The study, “Strain and rupture of HIV-1 capsids during uncoating,” was published March 2022 in the Proceedings of the National Academy of Sciences. The study authors are Elizabeth Lee, Alvin Yu, and Gregory Voth of the University of Chicago; John Briggs of the Max Planck Institute of Biochemistry; Barbie Ganser-Pornillos and Owen Pornillos of the University of Virginia.